タンパク質・化合物結合予測プログラムの実装
準備:RDkitをインストール
!pip install rdkit
% Total % Received % Xferd Average Speed Time Time Time Current
Dload Upload Total Spent Left Speed
0 0 0 0 0 0 0 0 --:--:-- --:--:-- --:--:-- 0
100 2415 100 2415 0 0 6822 0 --:--:-- --:--:-- --:--:-- 6822
add /root/miniconda/lib/python3.6/site-packages to PYTHONPATH
python version: 3.6.7
fetching installer from https://repo.continuum.io/miniconda/Miniconda3-latest-Linux-x86_64.sh
done
installing miniconda to /root/miniconda
done
installing rdkit
done
rdkit-2018.09.2 installation finished!
CPU times: user 334 ms, sys: 162 ms, total: 496 ms
Wall time: 2min 26s
題材:Tox21 Data Challenge 2014
前準備:Tox21データセットをダウンロード
!curl -Lo tox21_10k_all.sdf.zip https://bit.ly/2T6beHP
!unzip tox21_10k_all.sdf.zip
% Total % Received % Xferd Average Speed Time Time Time Current
Dload Upload Total Spent Left Speed
100 168 100 168 0 0 2210 0 --:--:-- --:--:-- --:--:-- 2210
100 2565k 0 2565k 0 0 1530k 0 --:--:-- 0:00:01 --:--:-- 1829k
Archive: tox21_10k_all.sdf.zip
inflating: tox21_10k_data_all.sdf
from rdkit import Chem
sup = Chem.SDMolSupplier('tox21_10k_data_all.sdf')
train = [mol for mol in sup
if mol is not None and 'NR-AR' in mol.GetPropsAsDict()]
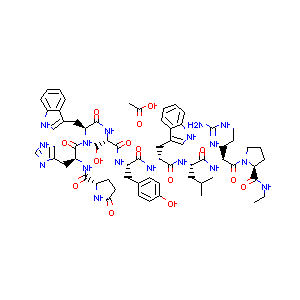
RDkitによる化合物構造式の表示
from rdkit.Chem import Draw
display(Draw.MolToImage(train[0]))
前処理:特徴抽出
Morgan Fingerprintによる特徴抽出
import numpy as np
from rdkit.Chem import AllChem
from rdkit import DataStructs
X = [ AllChem.GetMorganFingerprintAsBitVect(m, 2) for m in train]
X = [ np.asarray(x) for x in X]
X = np.vstack(X)
y = [m.GetIntProp('NR-AR') for m in train]
y = np.array(y)
print('X:', X.shape, 'y:', y.shape)
X: (9357, 2048) y: (9357,)
前処理:データの分割
トレーニングデータとテストデータに分割する。
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.25, random_state=1, stratify=y)
print('Labels counts in y:', np.bincount(y))
print('Labels counts in y_train:', np.bincount(y_train))
print('Labels counts in y_test:', np.bincount(y_test))
Labels counts in y: [8977 380]
Labels counts in y_train: [6732 285]
Labels counts in y_test: [2245 95]
学習:トレーニングデータによる予測モデル学習
パーセプトロンと呼ばれる学習アルゴリズムにより学習
from sklearn.linear_model import Perceptron
ppn = Perceptron(max_iter=40, eta0=0.1, tol=1e-3, random_state=1234)
ppn.fit(X_train , y_train)
Perceptron(alpha=0.0001, class_weight=None, early_stopping=False, eta0=0.1,
fit_intercept=True, max_iter=40, n_iter=None, n_iter_no_change=5,
n_jobs=None, penalty=None, random_state=1234, shuffle=True,
tol=0.001, validation_fraction=0.1, verbose=0, warm_start=False)
評価:テストデータによる精度評価
テストデータにおける精度を計算する。
from sklearn.metrics import accuracy_score, f1_score, precision_recall_fscore_support
y_test_pred = ppn.predict(X_test)
print("Acc:", accuracy_score(y_test , y_test_pred))
print("F:", f1_score(y_test, y_test_pred))
Acc: 0.9666666666666667
F: 0.5617977528089888
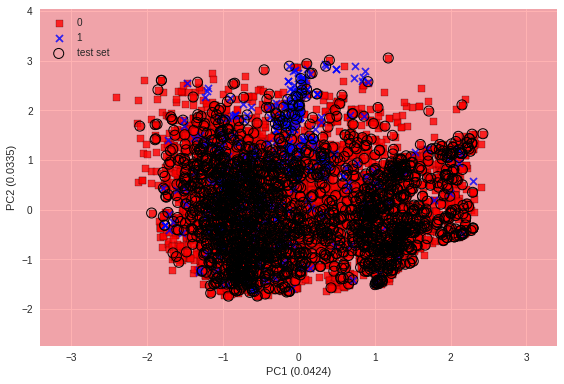
評価:予測モデルの可視化
主成分分析を用いて次元削減し、散布図にプロットする。
from matplotlib.colors import ListedColormap
import matplotlib.pyplot as plt
def plot_decision_regions(X, y, classifier, test_idx=None, pca=None, resolution=0.02):
# setup marker generator and color map
markers = ('s', 'x', 'o', '^', 'v')
colors = ('red', 'blue', 'lightgreen', 'gray', 'cyan')
cmap = ListedColormap(colors[:len(np.unique(y))])
# plot the decision surface
x1_min, x1_max = X[:, 0].min() - 1, X[:, 0].max() + 1
x2_min, x2_max = X[:, 1].min() - 1, X[:, 1].max() + 1
xx1, xx2 = np.meshgrid(np.arange(x1_min, x1_max, resolution),
np.arange(x2_min, x2_max, resolution))
x_inv = np.array([xx1.ravel(), xx2.ravel()]).T
if pca is not None:
x_inv = pca.inverse_transform(x_inv)
Z = classifier.predict(x_inv)
Z = Z.reshape(xx1.shape)
plt.contourf(xx1, xx2, Z, alpha=0.3, cmap=cmap)
plt.xlim(xx1.min(), xx1.max())
plt.ylim(xx2.min(), xx2.max())
for idx, cl in enumerate(np.unique(y)):
plt.scatter(x=X[y == cl, 0],
y=X[y == cl, 1],
alpha=0.8,
c=colors[idx],
marker=markers[idx],
label=cl,
edgecolor='black')
# highlight test samples
if test_idx:
# plot all samples
X_test, y_test = X[test_idx, :], y[test_idx]
plt.scatter(X_test[:, 0],
X_test[:, 1],
facecolor='none',
edgecolor='black',
alpha=1.0,
linewidth=1,
marker='o',
s=100,
label='test set')
from sklearn.decomposition import PCA
pca = PCA(n_components=2)
X_combined = np.vstack((X_train, X_test))
y_combined = np.hstack((y_train, y_test))
X_transformed = pca.fit_transform(X_combined)
test_idx = range(X_train.shape[0], X_combined.shape[0])
plot_decision_regions(X_transformed, y_combined, ppn, test_idx=test_idx, pca=pca)
plt.xlabel('PC1 ({:.4f})'.format(pca.explained_variance_ratio_[0]))
plt.ylabel('PC2 ({:.4f})'.format(pca.explained_variance_ratio_[1]))
plt.legend(loc='upper left')
plt.tight_layout()
plt.show()
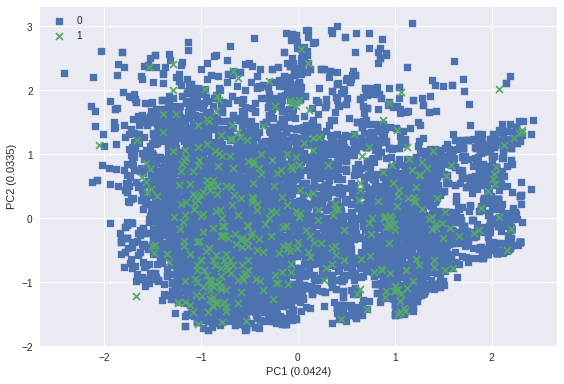
markers = ('s', 'x')
for idx, cl in enumerate(np.unique(y)):
plt.scatter(X_transformed[y==cl, 0], X_transformed[y==cl, 1],
marker=markers[idx], label=cl)
plt.xlabel('PC1 ({:.4f})'.format(pca.explained_variance_ratio_[0]))
plt.ylabel('PC2 ({:.4f})'.format(pca.explained_variance_ratio_[1]))
plt.legend(loc='upper left')
plt.tight_layout()
plt.show()
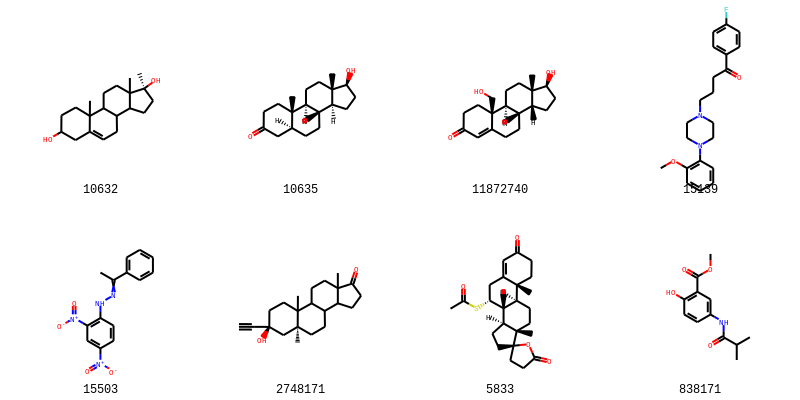
新規予測:候補化合物8種の結合予測
新しい化合物が結合するかを予測する。
!curl -Lo compound_list.sdf https://git.io/JvhPK
sup = Chem.SDMolSupplier('compound_list.sdf')
unknowns = [mol for mol in sup if mol is not None]
cids = [mol.GetProp('PUBCHEM_COMPOUND_CID') for mol in unknowns]
Draw.MolsToGridImage(unknowns, legends=cids, molsPerRow=4)
% Total % Received % Xferd Average Speed Time Time Time Current
Dload Upload Total Spent Left Speed
0 0 0 0 0 0 0 0 --:--:-- --:--:-- --:--:-- 0
100 53850 100 53850 0 0 60167 0 --:--:-- --:--:-- --:--:-- 60167
X_unknowns = [ AllChem.GetMorganFingerprintAsBitVect(m, 2)
for m in unknowns]
X_unknowns = [ np.asarray(x) for x in X_unknowns]
X_unknowns = np.vstack(X_unknowns)
y_unknowns_pred = ppn.predict(X_unknowns)
for cid, pred in zip(cids, y_unknowns_pred):
print(cid, pred)
10632 0
10635 1
11872740 1
15139 0
15503 0
2748171 1
5833 1
838171 0
ここで出力された機械学習による予測がAutodockによるドッキング解析結果とどのように異なるかを確認する。